
Protein Analysis and Characterization Services
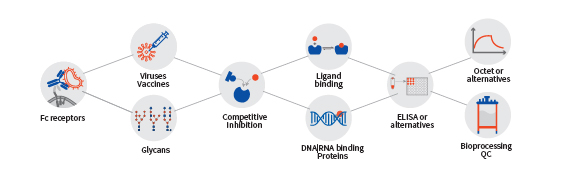
Aragen has set up an advanced and extensively equipped laboratory with experienced analytical scientists to provide a comprehensive characterization of novel biologics and biosimilars. We have a suite of analytical platforms to rigorously evaluate and assess your molecules using an array of biochemical and biophysical techniques. Whether you are interested in setting up assays, designing screens, confirming hits, or starting a developability study with your lead molecules, our expertise helps you move your project forward.
Comprehensive Protein Analysis and Characterization
Combining scientific expertise with expedited protocols to deliver target-specific antibody solutions.
Solutions for the stable expression and purification of novel molecules.
