 Back to News
Back to NewsTargeted Protein Degraders – The druggability perspective
November 04, 2023, By Satinder Singh, Pratima Srivastava
Targeted Protein degraders (TPDs) show promise in harnessing cellular machinery to eliminate disease-causing proteins, even those previously considered undruggable. Especially if protein turnover is low, targeted protein removal bestows lasting therapeutic effect over typical inhibition. The demonstrated safety and efficacy profile of clinical candidates has fueled the surge in the number of potential candidates across different therapeutic areas. As TPDs often do not comply with Lipinski’s rule of five, developing novel TPDs and unlocking their full potential requires overcoming solubility, permeability and oral bioavailability challenges. Tailored in-vitro assays are key to precise profiling and optimization, propelling breakthroughs in targeted protein degradation.
Graphical Abstract
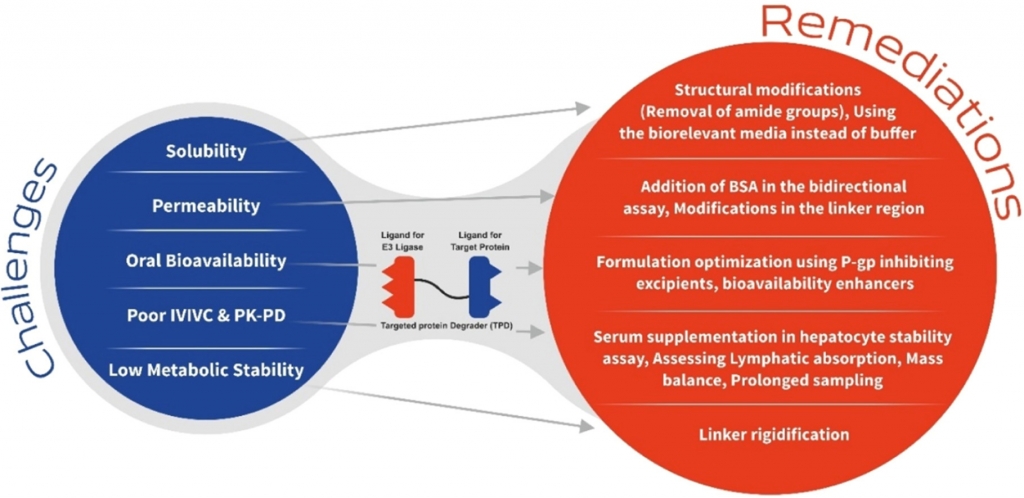
Introduction
TPDs are a class of macromolecular therapeutic compounds that harness the cell’s own protein degradation machinery to selectively break down proteins of interest mainly disease-causing1. In contrast to non-covalent inhibitors, TPDs offer a broader range of protein targets, including those previously regarded “undruggable” by conventional small molecules2. TPDs can be designed to degrade intracellular proteins, membrane proteins, and even transcription factors2,3.
One of the key advantages of TPDs is their high specificity. They capitalize on the binding affinity between the ligands present in the TPDs and sites present on target protein and the E3 ubiquitin ligase, ensuring precise degradation of the intended protein whilst evading unintended effects on other proteins4. Furthermore, compared to classical reversible inhibitors, TPDs exhibit a sustained and long-lasting impact on target protein levels due to their ability to induce protein degradation and removing it altogether5. This characteristic is particularly advantageous in chronic diseases where sustained protein suppression is necessary for therapeutic efficacy. TPDs also have the potential to overcome resistance mechanisms commonly observed with non-covalent inhibitors. By promoting protein degradation, instead of solely blocking protein activity, TPDs can bypass mutations or alterations that confer resistance to traditional inhibitors. Moreover, TPDs can be designed to simultaneously degrade multiple target proteins by incorporating multiple ligands into the same bifunctional molecule. This feature presents new possibilities for treating complex diseases involving multiple dysregulated proteins or pathways6.
In addition to their protein degradation capability, bifunctional TPDs can modulate protein function as well. These molecules can recruit specific proteins to facilitate protein-protein interactions or alter protein localization, thereby providing additional ways to influence cellular processes7.
Since 2001, when the first attempt to synthesize TPD (typical structure in figure 1) was made by Crews and Raymond Deshaies8, TPD chemistry and research has come a long way during last two decades.
TPDs made their clinical debut in 2018-19, and presently more than 10 oral and several parenteral TPDs are in Phase I/II clinical trials (Table 1).
Notably, ARV-110 has recently progressed into Phase III. TPDs created niche space of their own driven mainly by their potential to target undruggable proteins. The huge surge in number of pharma companies and institutes pursuing TPD programmes pushing them through preclinical to clinical stage are testimony to the potential of the approach in offering viable solutions for various diseases (Table 2).
Section snippets
The Degrader Landscape
The degrader landscape which conceptually evolved from MonoDAC (glue degraders) has now been extended from intracellular cytoplasmic proteins to extracellular or plasma membrane proteins as well, via employing different approaches like, ASGPR Targeting Chimeras (ASGPR-TCs), Bispecific Aptamer Chimera (BIAC), Antibody-based PROTAC (AbTAC), Autophagy-Targeting Chimera (AUTAC), Autophagosome-Tethering Compound (ATTEC), Chaperone Mediated Autophagy (CMA) based degrader and Lysosome-Targeting
Conclusion
Currently, several research groups are developing TPDs and tinkering with the proteome network to eliminate mutated or disease-causing proteins. It remains intriguing to see the long-term implications, if any, of wilfully engaging the proteasome machinery to completely remove the specific protein from the proteome web. The genome size of the western clawed frog Xenopus laevis, approximating 3.1 billion bases, is quite close in length to that of highly differentiated humans [3.2 billion bases]135
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Source – ScienceDirect
