

RNA-binding small molecules are gaining interest as therapeutics owing to RNA’s critical role in gene regulation and disease. However, their discovery requires precise characterization of RNA-ligand interactions, which is complicated by RNA’s inherent structural flexibility, chemical instability, and sensitivity to handling conditions. To meet these challenges, sensitive and reliable biophysical techniques are essential for maintaining RNA’s native conformation and accurately measuring binding events.
Surface Plasmon Resonance (SPR) offers a suitable biophysical technique that enables real-time, label-free analysis of RNA interactions with minimal sample consumption. However, successful application of SPR to RNA targets demands optimized assay conditions that preserve RNA stability and structure during immobilization and measurement. The primary challenges lie in maintaining biologically relevant folding, preventing degradation, and immobilizing RNA without compromising its functional integrity.
This case study illustrates how Aragen’s experts leveraged SPR to address key challenges in RNA-based SPR analysis, enabling precise kinetic profiling and identification of the most suitable high-affinity RNA-binding small molecule.
The experiment was performed using an advanced SPR platform. The laboratory was rigorously decontaminated to maintain an RNase-free environment, ensuring the integrity of RNA samples, and preventing degradation.
Multicycle kinetics (MCK) experiments successfully measured the binding affinities of various potential small molecules to the target RNA using the Biacore T200 platform. The team successfully utilized this approach to characterize a range of interactions, with the binding data for one of the most potent compounds presented in Figure 1, illustrating its detailed kinetics and binding affinity.
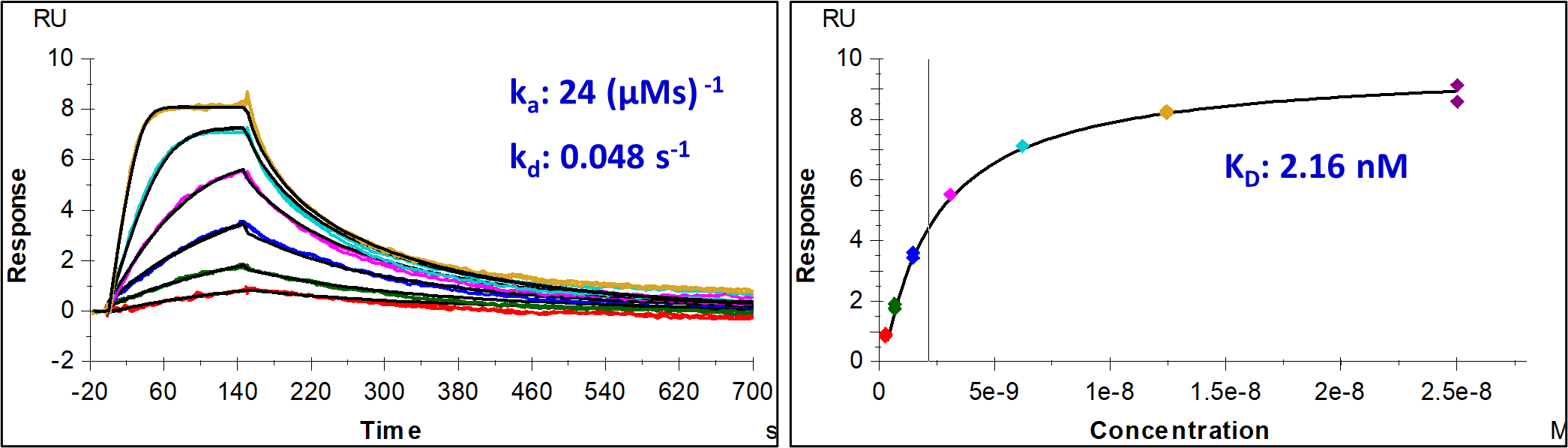
These results clearly demonstrate how Aragen’s SPR expertise, combined with experience in tailored handling of RNA-based samples, enables precise screening and detailed characterization of RNA-binding compounds. The approach applied herein is suitable to overcome the inherent challenges of RNA stability and folding, enabling more reliable and insightful RNA-targeted drug discovery at early stages.
With a decade of SPR expertise, we accelerate drug development by providing trusted insights that optimize drug candidate profiling and efficacy. We offer:
